A record-breaking beta barrel allows protein transport across the bacterial outer membrane
Research has uncovered an enormous transmembrane beta barrel structure through which the proteins move
New research from the Berks (Biochemistry) and Lea (Pathology) groups reveals how proteins are transported across the outer membrane of bacteria responsible for severe dental disease (peridontitis). This work, which is published in Nature [1,2], has uncovered an enormous transmembrane beta barrel structure through which the proteins move.
Movies. Electron microscopy density of the PorV (left) and Plug (right) Type 9 protein translocon complexes. The SprA protein is Rainbows coloured, partner proteins are grey. Specular density shows the position of the detergent micelle surrounding the translocon. The complexes are viewed in the same orientation as the Figure below. Movies courtesy of Susan Lea.
Pathogenic bacteria must be able to secrete proteins in order to manipulatetheir host organism. Major oral pathogens in the phylum Bacteroidetes, such as Porphyromonas gingivalis, export these proteins across two cell membranes. Protein transport across the outer membrane in these bacteria utilises the recently discovered Type IX Secretion System (T9SS).
All protein transport systems have to provide an aqueous pathway across the otherwise hydrophobic membrane bilayer. However, for the T9SS it was unknown which proteins build this protein conducting pathway, or translocon. A joint effort by Drs. Frédéric Lauber and Justin Deme working in the Berks and Lea groups has now identified the translocon and determined its structure in two mechanistically relevant states.
The team made the educated guess that a large T9SS component in the outer membrane with no sequence similarity to proteins of known function, called SprA, was likely to be the T9SS translocon. Imaging bacterial cells in which the native SprA protein was tagged with a fluorescent dye showed that there are only around ten copies of the protein in a cell. Nevertheless, the Oxford team was able to use a chromosomal tagging strategy to purify sufficient SprA from the native organism for structure determination by cryo-electron microscopy (cryo-EM) using the microscopes in the newly-established Central Oxford Structural Molecular Imaging Centre (COSMIC)[3]. Remarkably, SprA co-purified with three partner proteins. One of these proteins was an already-identified T9SS component named PorV. This protein is known to bind the T9SS substrate proteins at the cell surface following transport. However, the other two partner proteins turned out to be novel T9SS components. These proteins are a peptidyl-prolyl cis-trans isomerase (PPI) and a protein now called Plug. Sorting of the cryo-EM data revealed that the preparation actually contained two different types of complexes in which SprA is bound to either PorV or Plug, and the team was able to obtain structures for both complexes.
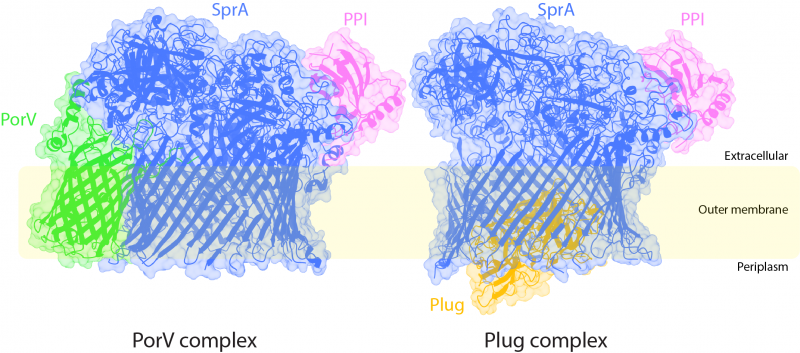
Figure. The two Type 9 translocon structures shown in ribbon representation inside the electron microscopy density. The inferred position of the bacterial outer membrane is indicated. The periplasm is the cellular compartment between the inner and outer membranes. The direction of protein transport is from the periplasm to the extracellular environment. The periplasmic end of the SprA barrel is open in the PorV complex but is occluded by the Plug protein in the Plug complex. Conversely, the lateral extracellular opening is blocked by the PorV protein in the PorV complex but accessible in the Plug complex. Image courtesy of Justin Deme.
Bacterial outer membrane proteins have a distinctive beta barrel architecture in which a sheet of polypeptide strands forms a membrane-spanning tubular structure surrounding a water-filled pore. In almost all cases the barrel is formed from a single protein chain. The cryo-EM structures of SprA reveals that it is also a single protein beta barrel, but of unprecedented size. The largest single protein outer membrane barrel protein previously known contains 26 polypeptide strands. However, SprA has 36 polypeptide strands, forming an internal pore that is two and a half times larger in cross sectional area than its nearest contender, and easily large enough to allow the passage of the proteins exported by the T9SS.
The SprA pore is capped on the extracellular end, but has a lateral opening to the external membrane surface. In the PorV-containing complex the lateral opening is occupied by PorV while the periplasmic end of SprA is accessible to substrate proteins. Conversely, in the Plug-containing complex the lateral opening is free but the interior end of the pore is occluded by the Plug protein. Binding of PorV and Plug to SprA is mutually exclusive due to changes in the shape of the SprA barrel between the two complexes.
Taken together the structural data suggests a novel protein transport mechanism in which there is alternating access to the SprA channel controlled by partner proteins, with the substrate protein exiting the external end of the channel as a complex with PorV.
Questions that now need to be answered include how the SprA translocon is able to recognise its transport substrates, how transport through the translocon is energised, and how the translocon interacts with other components of the T9SS.
Reference(s)
- Lauber, F., Deme, J.C, Lea, S.M., and Berks, B.C. (2018) Type 9 secretion system structures reveal a new protein transport mechanism (Article). Nature 564: 77–82 https://www.nature.com/articles/s41586-018-0693-y
- Blog: https://naturemicrobiologycommunity.nature.com/channels/346-behind-the-p...
- COSMIC: http://web.path.ox.ac.uk/~bioimaging/electronmicroscope/cryo_em.html
Ben Berks
19th November 2018




