Discovering the cellular RNA-binding proteins controlling virus infection
New research from Alfredo Castello’s laboratory published in Molecular Cell has discovered that virus infection rewires cellular RNA-binding proteins on a global level. This reflects two antagonistic processes: the virus hijacking key cellular resources and the antiviral defence mechanisms of the cell. This work provides a comprehensive snapshot of the virus-host cell battlefield and opens new avenues for the development of antiviral therapies.
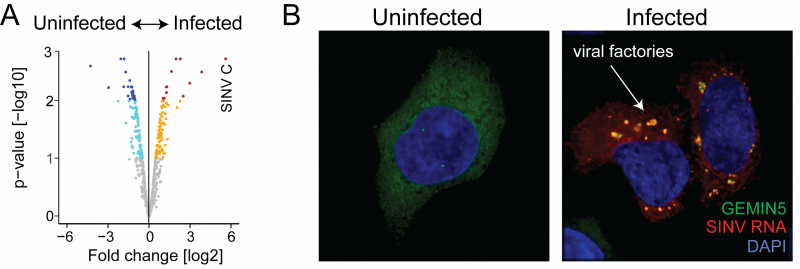
Figure: A) About two hundred human RNA-binding proteins (coloured dots) are regulated by the infection with SINV. B) RNA-binding proteins (in green) activated by SINV redistribute to the cellular compartments where the viral RNA accumulates (in red) (Click image to enlarge)
RNA-binding proteins are a critical component of the cellular machinery that dictate the fate of RNA molecules. As RNA virus genomes are small, they rely on host RNA-binding proteins to control the life of the viral RNA. However, which of these host proteins are required for virus infection remains largely unknown. Alfredo’s group developed a novel technique called ‘comparative RNA interactome capture’ to interrogate which RNA-binding proteins are involved in the infection of a model virus called Sindbis (SINV). This work uncovered that SINV infection alters the activity of more than 200 cellular RNA-binding proteins, thus rewiring cellular RNA metabolism (Figure 1).
The infection alters the activity of more than two hundred RNA-binding proteins – says Marko Noerenberg -. This is likely reflecting the battle between the virus and the host cell for the control of the cellular resources.
SINV turns off RNA-binding proteins participating in the nuclear life of the RNA, while activates key regulators of protein synthesis, RNA degradation and storage and in innate immunity. Removal of the proteins with enhanced activity from the cell SINV infection was severely affected, highlighting their key role at controlling viral replication. The intimate connection between these host proteins and the virus was confirmed by microscopy, showing that they accumulate at the places in the cell where the SINV replicates co-localising with the viral RNA (Figure 2).
Sindbis virus synthesises massive amounts of viral RNA – explained Manuel Garcia-Moreno -, and these RNA molecules act as spider webs that capture the proteins that the virus needs in the places where they are needed.
The researchers also reported a dramatic degradation of cellular RNAs while viral RNA accumulates. They discovered that cells lacking the exonuclease XRN1, mediator cellular RNA clearance, become resistant to SINV infection. Therefore, cellular RNA degradation emerges as a key process supporting viral infection. Moreover, their study revealed that the cellular protein GEMIN5 binds to the viral RNAs and inhibits their translation, which represents a new antiviral mechanism to control virus infection.
We discovered many RNA-binding proteins that are central for virus infection – added Alfredo Castello, who led this work-. These results open new avenues for the discovery of new therapies against viruses.
Reference
- Manuel Garcia-Moreno, Marko Noerenberg, Shuai Ni, Aino I. Järvelin, Esther González-Almela, Caroline E. Lenz, Marcel Bach-Pages, Victoria Cox, Rosario Avolio, Thomas Davis, Svenja Hester, Thibault J.M. Sohier, Bingnan Li, Gregory Heikel, Gracjan Michlewski, Miguel A. Sanz, Luis Carrasco, Emiliano P. Ricci, Vicent Pelechano, Ilan Davis, Bernd Fischer, Shabaz Mohammed and Alfredo Castello. System-wide profiling of RNA-binding proteins uncovers key regulators of virus infection. Molecular Cell. DOI: https://doi.org/10.1016/j.molcel.2019.01.017
Deepa Nath
21st February 2019




