How a group of conserved proteins orchestrate transcription termination in eukaryotes
A paper published in Nature Communications by Lidia Vasilieva's group in collaboration with researchers at the Division of Structural Biology at the University of Oxford (STRUBI) and the Max-Planck Institute for Biophysical Chemistry in Göttingen, Germany, sheds light on how a group of conserved proteins orchestrate transcription termination in eukaryotes (1).
Unexpectedly, their study also reveals that fission yeast, and possibly other eukaryotes, use a different mechanism from the model eukaryote budding yeast to regulate non-coding transcripts.
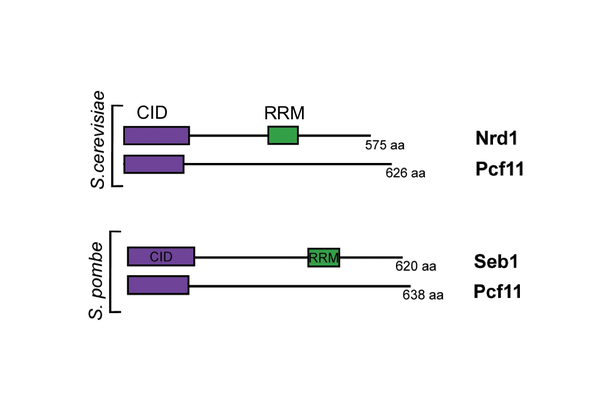
Figure 1: Homologous CID-containing proteins from budding yeast (S.cerevisiae) and fission yeast (S.pombe) with the conserved regions CID and RRM shown
All steps of transcription - initiation, elongation and termination - are tightly controlled. In eukaryotes, this control is directly coupled to RNA polymerase II (Pol II), a multi-protein complex whose largest subunit has a long C-terminal domain (CTD) with multiple repeats that are conserved across organisms. Many of the residues in the repeat can be reversibly phosphorylated and this is known to regulate key events during the transcription cycle. A conserved group of proteins required for transcription regulation are the CTD readers. These bind to phosphorylated residues on the CTD, in some cases via a conserved CTD-interacting domain (CID), but it is not fully understood how they mediate CTD function is regulating transcription.
Pol II makes both protein-coding and non-coding transcripts. These have very different functions in the cell and are regulated in different ways. Protein-coding transcripts leave the nucleus and move to the cytoplasm to be translated into proteins. In contrast, non-coding transcripts are primarily nuclear and are targeted by the cellular RNA degradation machinery for either 3'end processing or complete removal. In budding yeast, two essential CTD readers involved in transcription termination of protein-coding or non-coding transcripts, Pcf11 and Nrd1 respectively, are key to determining the different fate of these transcripts. Both have a CID, but Nrd1 recognises a different phosphorylated isoform of Pol II CTD from Pcf11 and also has an RNA recognition motif (RRM) that directly interacts with the RNA. How the CID proteins interact with Pol II, RNA and other factors determines which mechanism is used to terminate a particular transcript. Nrd1 also interacts with the cellular RNA degradation machinery, bringing it immediately to the transcription complex, and thereby helping to make functional non-coding RNAs and to degrade any unwanted transcripts.
Although CID-RRM proteins are very conserved, it is not known whether in other eukaryotes they are also involved in coupling regulation of non-coding and protein-coding transcripts to transcription termination. Lidia Vasilieva's group looked at how the CID-RRM and CID proteins from fission yeast - Seb1 and Pcf11 respectively (see Figure 1) - work to terminate transcripts. The group embarked on a series of structural and biochemical studies to find this out, with work largely carried out by D.Phil. student Sina Wittmann and assisted by another student, Beth Watts.
They looked first at where Seb1 binds during transcription. Surprisingly, they found Seb1 at the 3' end of both protein-coding and non-coding transcripts, suggesting that it might be functioning differently from Nrd1.
Figure 2: The structure of the Seb1 RNA-binding region, solved by X-ray crystallography and to 1.0 Å resolution. The canonical RRM domain is shown in blue and the second domain in green
The next set of experiments aimed to pin down the mechanism underlying Seb1's function. Collaborating with Jonathan Grimes and colleagues at STRUBI, the group used X-ray crystallography to understand how Seb1 binds to RNA. Solving the structure of Seb1 at very high resolution revealed two regions of the protein that are involved in RNA interactions - the RRM and a previously unknown second domain that showed an unusual arrangement. By using structural information, the group was able to show that RNA interacts with both domains. Introducing mutations that abolished Seb1 binding to RNA into living cells demonstrated that RNA binding was critical for the protein to function normally.
Turning their attention to the CID, the group found that this domain was also critical for Seb1 function but had different specificity to phosphorylated Pol II compared with Nrd1. When mutations were introduced into either the CID or RRM that abolished binding to Pol II and RNA respectively, this led to the production of excessively long RNAs on most Pol II-transcribed genes (protein-coding and non-coding). This defect was due to failure of Pol II to terminate properly, causing the enzyme to run into genes downstream and leading to a dramatic dis-regulation of transcription overall.
The results, says Dr Vasilieva, point to an unexpected role for Seb1 in fission yeast - that it is needed for termination of protein-coding transcripts as well as non-coding ones. This raises the question of how Pcf11 is contributing to transcription termination in this organism. 'Both Seb1 and Pcf11 are essential and not redundant, but both appear to be recruited to the same set of genes,' she comments. 'They must jointly enable termination to take place but in a different way from that seen in budding yeast.' The key to understanding their different roles, she adds, is identifying the different sets of proteins they interact with, work that will involve reconstructing the process in vitro.
The discovery that the termination pathways used for non-coding and protein-coding transcripts are the same in fission yeast is likely to hold true for other eukaryotes, including humans. But this leaves one question unanswered: what discriminates termination of non-coding and protein-coding transcripts in these organisms if it is not via the CID-RRM protein?' In budding yeast, Nrd1 brings the degradation machinery immediately to the complex,' says Dr Vasilieva. 'In contrast, Seb1 does not interact with that machinery so cannot be providing regulation of non-coding transcripts. It is needed as a factor to terminate non-coding transcripts, but something else must be providing this regulation.'
The work has provided a better understanding of how CTD-binding proteins bring about transcription termination, highlighting the importance of the RNA-binding region and of dynamic interactions the proteins are likely to have with other factors. Dr Vasilieva adds that their work also illustrates a key concept underlying much of biology. 'CTD-binding proteins are very important players in the termination process, connecting activities together. The concept that cellular machines must be recruited to the right place at the right time applies to any biological process.'
Reference
Wittmann S, Renner M, Watts BR, Adams O, Huseyin M, Baejen C, El Omari K, Kilchert C, Heo D-H, Kecman T, Cramer P, Grimes JM and Vasiljeva L. The conserved protein Seb1 drives transcription termination by binding RNA polymerase II and nascent RNA. Nat. Commun. 8, 14861 doi: 10.1038/ncomms14861 (2017)
Jane Itzaki
3rd April 2017




