How displaceable are water molecules?
New Research by the Biggin Group
How displaceable are water molecules?
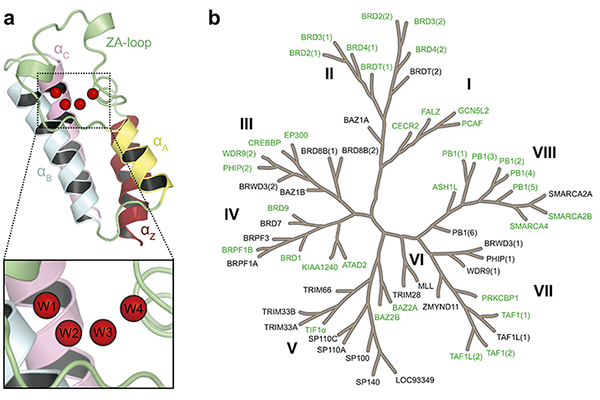
Figure: (a) Structure of the bromodomain fold and its conserved water network. (b) Phylogenetic tree of the human bromodomain family, with the 35 bromodomains considered in our study highlighted in green
The Biggin group, in collaboration with colleagues from the University of Southampton and industry, has just published a large-scale prediction of water-stability in a family of proteins that are key targets for the treatment of many different cancers. Being able to design compounds that displace these key waters is a common strategy in medicinal chemistry, but knowing which waters to target or if they are even likely to be displaceable at all is an extremely challenging problem. The group employ a method, recently developed by the Essex group in Southampton, called Grand Canonical Monte Carlo (GCMC) to make predictions about which water molecules in bromodomains are likely to be displaceable.
Bromodomains are small protein modules that recognise acetylated lysine on histones and thereby regulate gene expression by recruiting various transcription factors. To date, 61 bromodomains have been found in 46 different human proteins. As such, they are being investigated as a promising epigenetic target for diseases like cancer and inflammation. A few bromodomain inhibitors are in fact already in clinical trials for the treatment of leukaemia, midline carcinoma, and progressive lymphoma. Because of this, there is great interest in further exploring the therapeutic potential of this protein family.
Although there is considerable sequence diversity, bromodomains share very similar binding pockets, which makes the discovery of selective probes challenging. Bromodomains are amenable to study by X-ray crystallography and structures are available for many different types of bromodomain (see Figure 1). The structures revealed a conserved water network formed by four water molecules at the base of the binding site (Figure 1) and it is these waters which, if displaceable, may offer opportunities to gain affinity and/or improve selectivity.
The problem is that the rational identification of displaceable waters molecules is almost impossible by simple inspection of crystal structures, and even more difficult is a quantitative estimation of their stability. In fact, the stability of water molecules in protein cavities cannot be determined experimentally, as water is also the solvent that proteins reside in. Aside from pure trial and error, the only realistic option is to employ computational approaches.
Approaches that use all-atom statistical mechanics-based simulations, like Monte Carlo or molecular dynamics are the most suitable approaches to study water stability in part because they inherently deal with the problem of entropy. The GCMC method allows us to investigate the arrangement of possible water networks at a moderate computational cost and thus allowed us to study all 35 bromodomains with resolved X-ray structures using three different water models. The study published today in Communications Chemistry presents the results of our calculations where we quantify the stability of the four conserved water molecules in bromodomains in terms of their binding free energy to the protein.
Although it is difficult to make a direct comparison to experiment, our calculations make predictions that are in excellent agreement with drug-design programs that have successfully made compounds that displace waters in certain bromodomains. This gives us confidence that the predictions across this whole family of proteins will provide useful information for future drug-design projects and in particular may help in the prioritization of inhibitor design.
Associated Links:
Philip Biggin
29th March 2018




