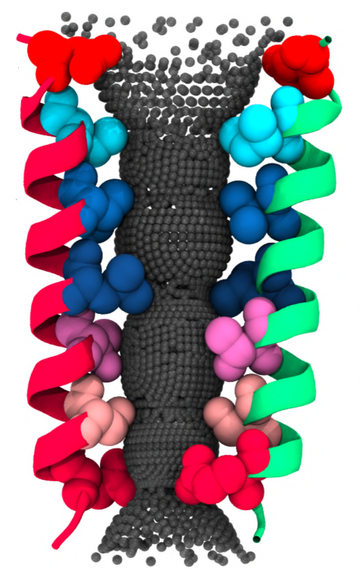
Figure 1. The pore lining structure of the serotonin receptor
The work was carried out in Prof Mark Sansom's laboratory in collaboration with Prof Stephen Tucker at the Department of Physics in Oxford, and has been published in Structure [1].
Ion channels act as pores that allow the movement of ions across the cell membrane. They play key roles in fundamental physiological processes and are especially important in electrically excitable cells within the nervous system. Ion channels are highly dynamic and can undergo conformational transitions as they move between a closed state that is impermeable to ions and an open state that allows the flow of ions. Although recent advances in structural biology are yielding an increasing array of high resolution structures, a major limitation is that the functional state of a channel (i.e., open versus closed) is often unclear. Better tools are needed to annotate the functional state of a channel.
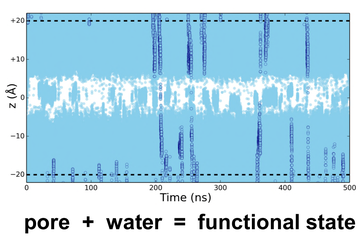
Figure 2. A simulated trajectory of water molecules crossing the central hydrophobic barrier in the pore
Several ion channels exhibit a property referred to as 'hydrophobic gating'. When water and ions are present within narrow hydrophobic pores they exhibit unusual behaviour. Unfavourable interactions between water and the lining of a hydrophobic pore, promotes a 'dewetted' state that is devoid of water. Expulsion of water presents an energetic barrier to ion permeation. The ability of a pore to become hydrated therefore represents a reliable indicator of permeability.
Using this principle, Mark Sansom and colleagues, carried out three successive simulations on the serotonin receptor (5-HT3R) structure. All three approaches correctly predicted the functionally closed state of the receptor. This method has since been extended to glycine receptors and other families of ion channels.
This study demonstrates that molecular dynamics simulations can be used to accurately annotate ion channels based on the principles of hydrophobic gating. The next step will be to provide an automated web-accessible tool to help with annotation of new ion channel structures that are now beginning to emerge.
Trick, J.L., Chelvaniththilan, S., Klesse, S., Aryal, P., Wallace, E.J., Tucker, S.J., Sansom, M.S.P. (2016) Functional Annotation of Ion Channel Structures by Molecular Simulation. Structure 24(12): 2207-2216
Deepa Nath
25th January 2017