New paper shows how transcription terminates with the help of a phosphatase
New research from Lidia Vasilieva's lab sheds light on how transcription termination in eukaryotes is controlled.

Vasilieva Lab members: Tea Kecman, Adrien Birot, Lidia Vasilieva, Krzysztof Kus and Dong-Hyuk Heo
The findings, from Dr Vasilieva's group in collaboration with colleagues in the Department of Chemistry, are published in Cell Reports (1) and demonstrate that conserved mechanisms are used at different steps of transcription. The work will help to
fill the gaps in our understanding of how RNA polymerase II is released at the end of transcription.
All three stages of the transcription cycle of mRNAs - initiation, elongation and termination - are tightly regulated. The multi-protein complex RNA polymerase II (Pol II) interacts with a different set of factors to regulate the transition between
these stages. The factors modulate how Pol II behaves, changing its ability to interact with DNA, incorporate nucleotides or pause.
Pol II interacts with factors via its core and also via the C-terminal domain (CTD) of its largest subunit, which consists of heptad repeats in which a number of residues can be reversibly phosphorylated during the transcription cycle. At the initiation
to elongation transition, initiation factors that are bound to the core are replaced by elongation factors, causing Pol II to pause. Termination is essential for 3' end formation of functional mRNA, mRNA release, and Pol II recycling, but the
changes that take place as Pol II moves from elongation to termination are less well understood. In fission yeast, an organism that serves as a good model for more complex eukaryotes, a conserved termination factor called Seb1 is known to interact
directly with the CTD and is essential for regulating 3' end processing and termination.
DPhil student Tea Kecman, postdoc Krzysztof Kus and their colleagues in Dr Vasilieva's group set out to explore how the transcription machinery is controlled during termination in fission yeast. They wanted to investigate further how phosphorylation
of the CTD controls the late stages of transcription. The researchers looked at the CTD phosphorylation pattern at gene 3' ends across the genome and found that phosphorylation of Thr4 on the CTD was enriched. The level of Thr4 phosphorylation
was regulated by the conserved phosphatase PP1. When PP1 was deleted, transcription termination was disrupted, suggesting that dephosphorylation of Thr4 is important for termination.
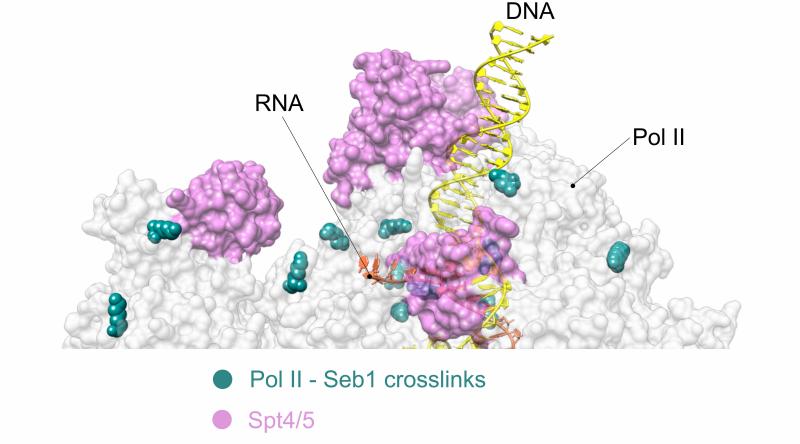
Binding surfaces of elongation and termination factors on Pol II overlap. Pol II crystal structure with Spt4/5 (PDB ID: 5XON ) (pink) showing mapped positions of lysine-lysine cross-links (turquoise spheres) between Pol II subunits (grey) and termination factor Seb1
The researchers also found that when PP1 was absent, Seb1 could not bind to Pol II. To investigate how lack of PP1 interferes with Seb1/Pol II interactions, they reconstituted Seb1/Pol II complex from purified Seb1 and native Pol II. Collaborating
with in the Department of Chemistry, they carried out cross-linking coupled to mass spectrometry analysis of the complex. They were surprised to find that Seb1 interacts directly with the core of Pol II and that binding mapped to a region where
an elongation factor called Spt5 binds. The results suggested that factor exchange takes place during the transition from elongation to termination as well as from initiation to elongation. By carrying out further experiments, the researchers
showed that Seb 1 recruitment is dependent on dephosphorylation of Spt5 by PP1.
The group's findings point to a complex level of control in which both Thr4 and termination factor exchange are regulated by PP1, implicating PP1 as a master regulator of the elongation to termination transition. This is analogous to the transition
from initiation to elongation, which is regulated by a kinase that phosphorylates both CTD and also Pol II interacting factors.

Model proposed. Conserved PP1 orchestrates transition from elongation to termination by dephosphorylating Spt5 and CTD-Thr4P at the end of the transcription cycle
Having discovered how Seb1 binds to Pol II to bring about termination, the researchers now have an opportunity to probe the mechanism by which Pol II is dislodged from DNA. Dr Vasilieva suggests that Seb 1 may alter Pol II in such a way that comes
off the DNA.
'Seb1 is an essential factor that mediates 3' end processing and coupling to termination. It's not just bridging physically two molecular machines but also transferring information; it's perhaps a bit like a molecular neuron. Understanding how Seb
1 dislodges Pol II from DNA is a very important step towards understanding eukaryotic transcription termination. We will need to use a combination of multidisciplinary approaches to achieve this.'
The group has recently developed an in vitro system, which will be an important tool alongside other approaches such as genetics to understand what is happening. The role of CTD Thr4 dephosphorylation by PP1 in regulating termination is also
unknown and is another piece of the puzzle that remains to be solved.
As more and more of details of eukaryotic transcription are revealed through studies such as this one, the sheer complexity of the process - involving the orchestration of multiple protein complexes, kinases and phosphatases - is impressive by any
standards.
Reference
- Kecman T., Kuś K., Heo DH., Duckett K., Birot A., Liberatori S., Mohammed S., Geis-Asteggiante L., Robinson C. and Vasiljeva L. (2018). Elongation/Termination Factor Exchange Mediated by PP1 Phosphatase Orchestrates Transcription Termination.
Cell Reports. 25. 259-269.e5. https://doi.org/10.1016/j.celrep.2018.09.007
Jane Itzhaki
5th November 2018




