Protein antibiotic hijacks iron transporter to kill bacterial cells
Colin Kleanthous' lab has shown for the first time how a toxin released by bacteria delivers its toxic payload into cells.
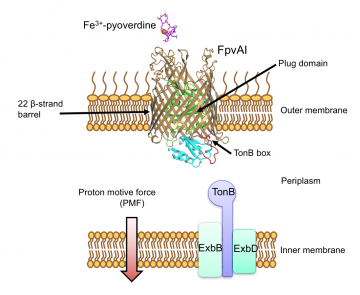
FpvA1 transports iron as a complex, Fe-pyoverdine, across the bacterial outer membrane by coupling to the Proton Motive Force (PMF) via the inner membrane protein TonB
In a recent PNAS paper, the group demonstrates that the bacteriocin hijacks a transporter normally used for bringing iron into the cell, to sneak across the outer membrane (1). The work will help on-going efforts to develop bacteriocins, which are naturally occurring protein antimicrobials, as next-generation antibiotics.
Bacteriocins are produced by bacteria to kill their neighbours during competition for resources. They kill cells through a variety of ways, most often by a protein domain that cleaves nucleic acids in the cytoplasm. Gram-negative bacteria are resistant to several classes of antibiotics because their outer membrane excludes these molecules, contributing to multidrug resistance. Yet the outer membrane is readily bypassed by bacteriocins, which are large, folded proteins. As bacteria do not have recognised protein import systems, it has been a mystery how bacteriocins navigate their way into bacterial cells.
Now work from the Kleanthous lab In this work, carried out largely by Professor Kleanthous' former DPhil student, Paul White, in collaboration with departmental colleagues Christina Redfield and Shabaz Mohammed and collaborators in the Biochemistry Department and at the University of Glasgow has revealed that the bacteriocin pyocin S2 (pyoS2) translocates across the outer membrane in the pathogenic bacterium Pseudomonas aeruginosa by mimicking the import of iron through its transporter, FpvA1. PyoS2 was known to require FpvAI as a receptor but nothing was known about how the bacteriocin entered the cell. Since pyoS2 is effective in treating P. aeruginosa -induced pneumonia in mouse, the work offers hope that such molecules could be developed as antibiotics.
P. aeruginosa , like other pathogens, must acquire iron once inside its host. It does so by releasing iron-capturing siderophores that chelate iron from the environment, which are then imported back into the cell where the iron is stripped out. FpvA1 recognises the siderophore ferripyoverdine (Fe-Pvd). Fe-Pvd binds to FpvA1 in the outer membrane and is transported through the receptor in conjunction with the inner membrane protein TonB, which provides the energy needed for transport. Energy is required because FpvA1 is a barrel-like protein that has a large central channel sealed with a protein plug. This plug domain stops molecules leaking in and out of the cell but also impedes Fe-Pvd passing through. TonB, in conjunction with the PMF (proton motive force), provides the energy needed to pull part of the plug out of FpvA1, opening a hole big enough for the siderophore.
In this work, carried out largely by Professor Kleanthous' former DPhil student, Paul White, in collaboration with departmental colleagues Christina Redfield and Shabaz Mohammed and others, a wide range of techniques were used to explore how pyoS2 recruits FpvA1 to get across the outer membrane.
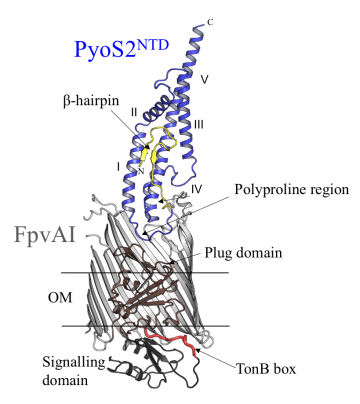
Crystal structure of the complex of FpvA1 and the N-terminal domain of pyoS2 (PyoS2NTD) showing pyoS2’s polyproline region, which mimics Fe-Pvd, and its TonB box, housed within the beta-hairpin
The first step was to use X-ray crystallography to solve the structure of the N-terminal domain of pyoS2 and, together with biophysical measurements, show how this region binds to FpvA1 and exploits it as a translocator. These experiments revealed that the N-terminal domain has a polyproline region that mimics the shape of Fe-Pvd, inducing the same conformational changes within the plug as those triggered by Fe-Pvd. 'The proline-rich region kinks the polypeptide chain so that it looks a bit like the natural ligand,' explains Professor Kleanthous. 'This region is found in several pyocins, so it seems that the mimicking system applies to these as well.'
From there, the group used fluorescence microscopy to follow how the N-terminal domain is translocated in live P. aeruginosa cells and probe the influence of TonB and the PMF. They identified a putative TonB binding site within the N-terminal domain and showed that this recruits TonB to allow translocation of pyoS2. Translocation requires energy because bacteriocins are folded, which is important for them to recognise the correct receptor. By developing a GFP-based cross-linking method in which they could trap pyoS2 within FpvA1, the group went on to identify which residues in the pyocin and the receptor make contact with each other as the bacteriocin translocates through the channel.
The findings from these experiments indicated that pyoS2 exploits the same mechanism as Fe-Pvd to open up the receptor, tricking it into delivering a big protein rather than a small molecule. Whilst the group was able to deduce that the N-terminal domain must unfold in order to be translocated through FpvA1, they do not yet know what induces the TonB binding region to go through the channel nor how TonB exerts its unfolding force on the pyocin.
The results show that pyocin - and quite possibly other bacteriocins that use TonB - have found clever ways of getting into cells. 'It's remarkable that pyoS2 appears to trick FpvA1 into thinking that it's a siderophore being brought into the cell,' says Professor Kleanthous.
He adds that there are many different directions the work could follow. 'We've solved the X-ray structure of the N-terminal domain of pyoS2 but we don't know how the other domains move through the pore. We'd also like to find out what induces the TonB binding sequence of the pyocin into the hole and how the cytotoxic endonuclease gets into the cytoplasm.' As well as this, the group would like to understand more about the process to determine what the scope for piggybacking into bacteria is. 'By understanding the process better, we can ask whether we can import other types of molecules, maybe even other types of toxic domains.'
The work will also be important for studies that Professor Kleanthous and colleagues are pursuing to explore whether bacteriocins might be developed as next-generation antibiotics. 'Bacteriocins are big proteins, unlike antibiotics that are small organic molecules, so they would represent a completely different form of antimicrobial therapy.' The approach will help to tackle the worldwide problem of multidrug-resistant bacteria.
References
- Exploitation of an iron transporter for bacterial protein antibiotic import. White P., Joshi A., Rassam P., Housden N.G., Kaminska R., Goult J.D., Redfield C., McCaughey L.C., Walker D., Mohammed S. and Kleanthous, C. PNAS 2017 doi: 10.1073/pnas.1713741114
Jane Itzhaki
1st November 2017




