The final step in bacterial lipoprotein maturation
Simulations and molecular mechanisms of apolipoprotein N-acyl transferase (Lnt).
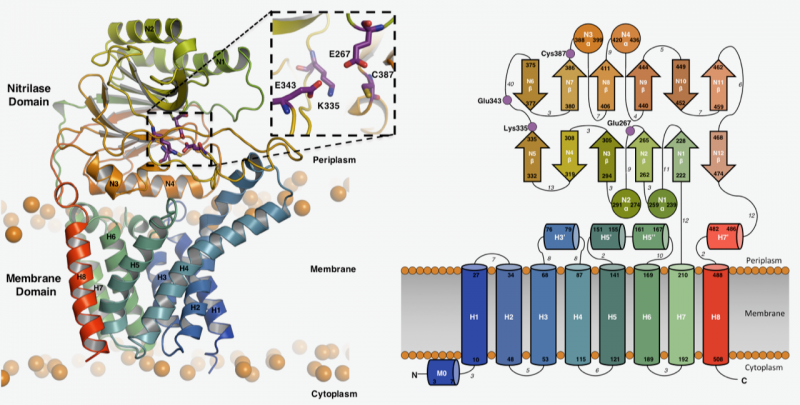
Figure 1: Structure and topology of Lnt
Gram-negative bacteria express numerous lipoproteins on their outer membrane. These proteins serve vital roles in bacteria and contribute to pathogenicity and antibiotic resistance. Understanding how lipoproteins are made will pave the way to develop new antibiotics.
Lipoproteins are synthesized in a precursor form which undergoes further post-translational modifications to form a mature protein. Three key enzymes, Lgt, LspA and Lnt, act sequentially to attach lipids to these proteins. These enzymes are present in many disease-causing bacteria, but are absent in humans, thus making them valuable drug targets.
Dr Phillip Stansfeld, in collaboration with the group of Prof Martin Caffrey at Trinity College, Dublin, recently solved and simulated the structure of LspA, the second enzyme in the lipoprotein maturation pathway (1). They have now, in association with the groups of Dr Meitian Wang and Prof Nienke Buddelmeijer, published details of the third and final enzyme, Lnt, in Nature Communications (2).
Precursor forms of lipoproteins are initially inserted and anchored in the bacterial inner membrane by a membrane-spanning helix. Here, the first processing enzyme, Lgt, attaches a diacyl lipid to a highly-conserved cysteine. As a result of this reaction, the protein is secured in the membrane by the membrane spanning helix and the lipid. The second enzyme, LspA, then removes the membrane-spanning helix, leaving the lipoprotein attached to the inner membrane by only the diacyl lipid tail. Some lipoproteins are not modified any further, while the rest are acted upon by the third and last enzyme, Lnt. Lnt adds another lipid to the diacyl lipid tail to form the triacylated and mature lipoprotein. The mature form is then transported to the outer membrane of gram negative bacteria where it can carry out its function; e.g. LptDE (3) and BAM (4).
This article reports the crystal structures of Lnt from two common disease-causing bacteria, Pseudomonas aeruginosa and, Escherichia coli (Fig 1). The structures reveal that the Lnt enzyme from both bacterial species, adopts a similar three-dimensional architecture. Lnt enzymes have a cylindrically-shaped membrane domain consisting of eight transmembrane helices. On top of this large domain sits a globular domain which projects into the periplasmic space.
The active site of Lnt lies between these two domains and sits slightly above the inner membrane, where it is poised to interact with lipoprotein substrates. The structures are consistent with the proposed two-step ping-pong reaction mechanism.
Molecular simulations were performed for all proposed states within the reaction (Fig 2) and suggest plausible routes by which lipid and lipoprotein substrates and products enter and leave the active site. The crystal structures, in conjunction with the molecular mechanisms and dynamics should facilitate rational design of selective inhibitors of this reaction, and therefore the development of novel antibiotics.
Reference(s)
- LspA - the second lipoprotein processing enzyme.
http://science.sciencemag.org/content/351/6275/876 - Lnt - the third and final maturation enzyme for lipoproteins.
https://www.nature.com/articles/ncomms15952 - BAM - responsible for inserting proteins into the bacterial outer membrane:
http://www.nature.com/nature/journal/vaop/ncurrent/abs/nature17199.html - LptDE - the protein that constructs the bacterial outer membrane:
http://www.nature.com/nature/journal/v511/n7507/full/nature13464.html
Phill Stansfeld and Deepa Nath
4th July 2017




