Kleanthous Group
Welcome to the Kleanthous Research Group
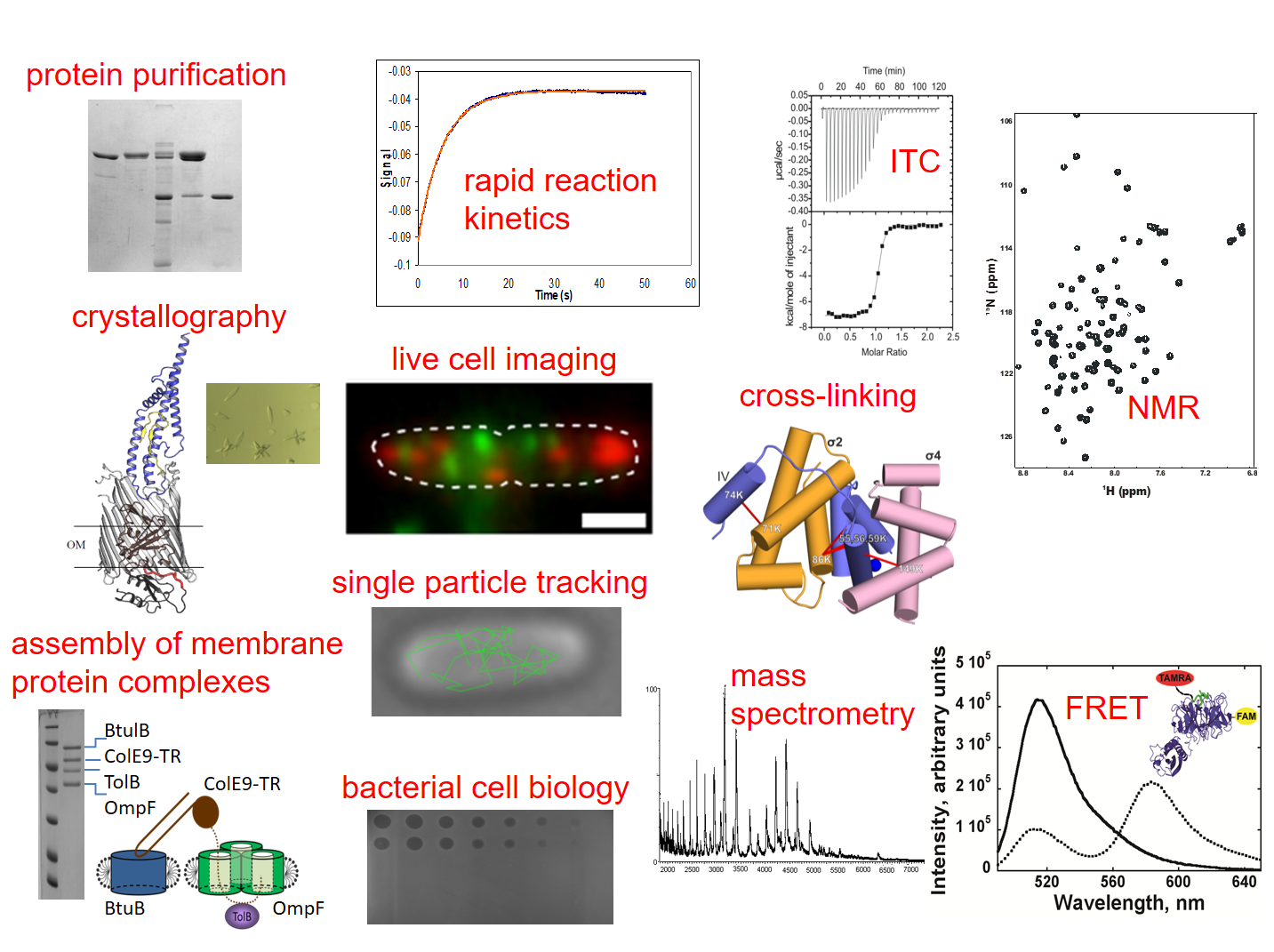
Protein-protein interactions in the Gram-negative bacterial cell envelope
In my laboratory, we aim to understand how protein-protein interactions in bacteria help support the organisation, structure and stability of the outer membrane and how such interactions can also be exploited by protein antibiotics known as bacteriocins to kill bacterial cells. Our research typically incorporates a range of biochemical, biophysical, structural and cell-based techniques, as summarised in the figure above. Our work is supported by the Wellcome Trust, ERC, BBSRC and MRC.
Awards and Distinctions
2018 Elected member of EMBO
2012 Iveagh Chair of Microbial Biochemistry, Oxford
2011-2013 Chair, The Biochemical Society
Bacteriocin translocation – Bacteriocins are species-specific protein antibiotics that parasitize a variety of outer membrane and periplasmic proteins in Gram-negative bacteria. Our work centres on the entry mechanisms of colicins, which target Escherichia coli, pyocins, which target Pseudomonas aeruginosa, klebicins, which target Klebsiella pneumoniae, and marcescins, which target Serratia marcescens. These toxins serve as important agents of competition within microbial communities. We study both nuclease (DNases, rRNases and tRNases) and pore-forming bacteriocins, which use their network of protein-protein interactions within the cell envelope to establish a translocon complex that delivers a toxic domain into the cell. Hence, bacteriocin translocation represents a highly simplified model for understanding cellular protein import.
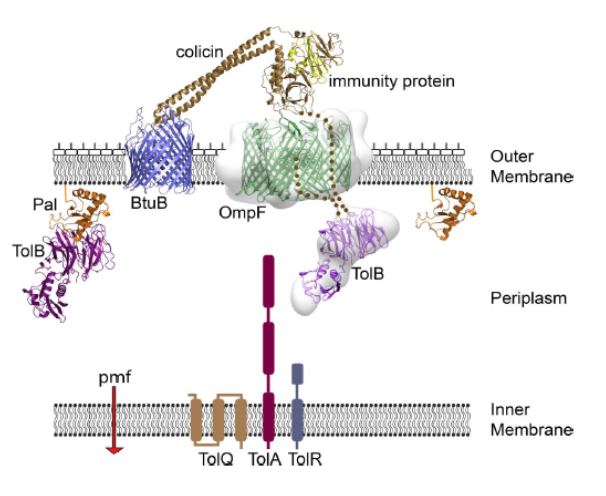
Colicin E9 delivers a linear peptide signal through the pores of the Escherichia coli porin OmpF in the outer membrane to capture the energized TolQRAB complex in the inner membrane and periplasm. The colicin has two OmpF-binding sites within its 83-residue intrinsically unstructured translocation domain (dotted line). These allow the colicin to capture TolB on the other side of the membrane by threading this unstructured domain through two pores of an OmpF trimer, one going into the cell the other coming back out again. OmpF is normally used by the bacterium to exchange metabolites with the environment and bring in nutrients such as glucose into the cell. This intricate cascade of protein-protein interactions initiates entry of the colicin’s nuclease killing domain into the cell. See Housden et al (2013) Science 340, 1570 for further details.
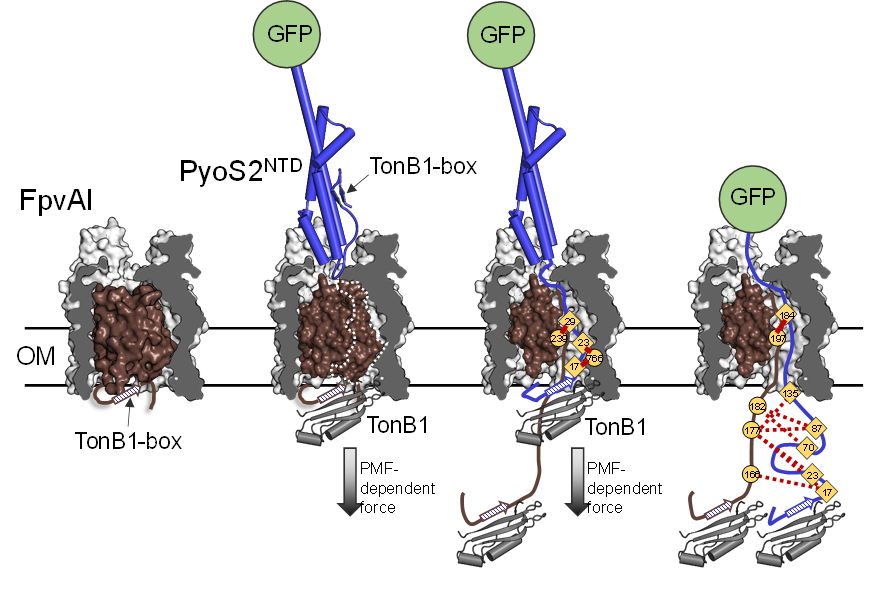
Pyocin S2 tricks the iron uptake receptor FpvAI into importing it across the Pseudomonas aeruginosa outer membrane. The N-terminal domain (NTD) of pyocin S2 (a nuclease bacteriocin) binds to the FpvAI receptor inducing a force-dependent conformational change, which opens the receptor. Pyocin S2 then delivers a signal (the TonB1 box) that binds TonB1 in the periplasm, inducing a further force-dependent step that unfolds the toxin and brings it into the cell. Green fluorescent protein (GFP) was used here to block entry. This approach allowed us to trap this partially translocated state and map its protein-protein interactions by photoactivated crosslinking (red and dotted lines). See White et al (2017) PNAS 114, 12051 for further details.
Developing bacteriocins as protein antibiotics – The rise of multidrug resistant bacteria coupled with the lack of new classes of antibiotics in over 30 years means there is a pressing and urgent need for new antibiotics especially for molecules that target pathogenic Gram-negative bacteria. In collaboration with groups in Oxford, Glasgow, Cambridge and UCL, we are exploiting the species selectivity and potency of bacteriocins as antimicrobials for Pseudomonas aeruginosa and Klebsiella pneumoniae, which are major causes of opportunistic and hospital-acquired infections world-wide.
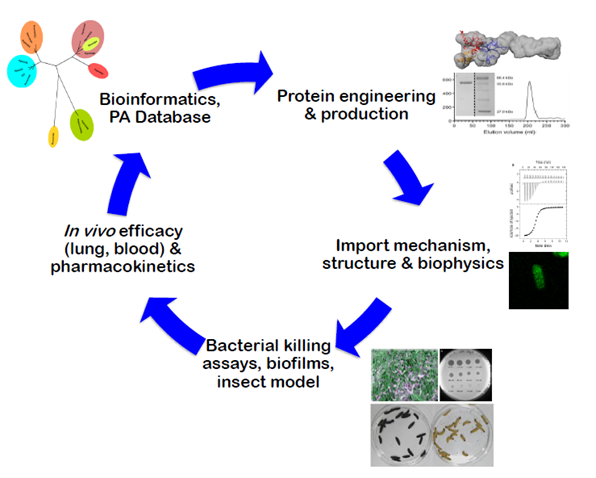
Pipeline being used to discover, characterise and develop protein antibiotics (bacteriocins) that target lung and blood infections.
Supramolecular assembly of outer membrane proteins – The asymmetric bilayer of the outer membrane contains outer membrane proteins (OMPs) and lipoproteins. Using colicins as OMP-specific probes, we recently discovered that OMPs cluster to form ‘OMP islands’ in the membrane and that this clustering behaviour underpins their turnover. We also demonstrated that the clustering behaviour of OMPs could be replicated in vitro using supported bilayers. Through the creation of a variety of OMP-specific labels we are dissecting the organisation of OMP islands and investigating the ramifications of these micro-domains to bacterial physiology. For example, we demonstrated recently that the inherent organisation of outer membrane proteins into islands becomes imprinted on inner membrane proteins when they are connected by an energized protein bridge.
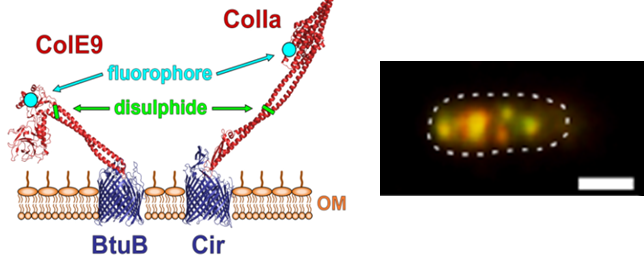
Outer membrane proteins (OMPs) co-localize in supramolecular assemblies in the outer membrane of E. coli. Left-hand panel, engineering colicins E9 and Ia as specific, high affinity labels of the OMPs BtuB and Cir. BtuB is the transporter for vitamin B12 while Cir is a siderophore transporter. The two colicins were inactivated by internal disulphide bonds prior to labelling with fluorophores. Right-hand panel, E. coli OMP islands imaged by TIRF microscopy and showing the co-localization of BtuB (labelled with ColE9-AF488, green) and Cir (labelled with ColIa-TMR, red). Scale bar is 1 mm. See Rassam et al (2015) Nature 523, 333 for details.
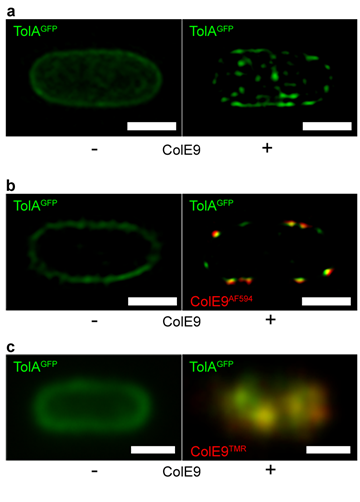
BtuB-bound ColE9 induces clustering in the inner membrane protein TolA. a, 3D-Structured illumination microscopy (SIM) images of E. coli cells expressing GFP-TolA showing how formation of a transenvelope bridge induces clustering of TolA in the inner membrane. b, 2D-SIM z-slice showing significant co-clustering (yellow fluorescence) of GFP-TolA and ColE9AF594 in the inner and outer membranes, respectively. c, TIRFM data showing co-localization of GFP-TolA clusters in the inner membrane and OMP-bound ColE9TMR islands (yellow fluorescence). See Rassam et al (2018) Nat Commun 9, 1082 for details.
The Tol-Pal assembly – Tol-Pal is a little understood complex that is required for the stable maintenance of the Gram-negative outer membrane and which is recruited to the cell division apparatus late during division. We are trying to uncover the native function of the Tol-Pal assembly, how and why it is coupled to the proton-motive force across the inner membrane and the mechanism by which bacteriocins subvert the assembly to initiate import across the outer membrane.
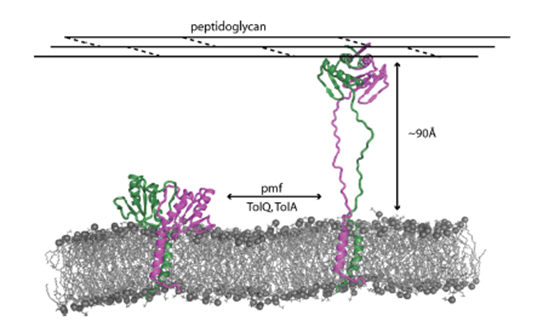
Postulated structural transition for the proton motive force (PFM) linked TolR protein. TolR is a dimeric protein that is part of the TolQRA complex in the inner membrane of Gram-negative bacteria. This work reported the crystal structure of a strand-swapped dimer of TolR (left hand figure). We postulated that strand unwinding would allow TolR to contact the peptidoglycan cell wall 90 Å away (right hand figure), which is a requirement for function of the Tol-Pal assembly. These transitions are thought to link to Tol-Pal stabilisation of the outer membrane of Gram-negative bacteria. See Wojdyla et al (2015) J. Biol. Chem. 290, 26675 for further details.
Kleanthous lab publications (Pubmed)
External appointments, society memberships and conference organisation
-
2016-2018 Research Council of Norway. Medicine Health Sciences and Biology panel.
-
2016-present Editor-in-chief, Emerging Topics in Life Sciences, Portland Press
-
2014-present John Innes Centre Science & Impact Advisory Board
-
2013 Organising Committee ESF-EMBO conference on 'Molecular Perspectives on Protein-Protein Interactions', Polonia Castle, Pultusk, Poland
-
2012 Organising Committee, Biochemical Society Focus Meeting 'How bugs kills bugs: Progress and challenges in bacteriocin research', University of Nottingham.
-
2011-2012 Chair, Wellcome Trust Basic Science Interview Committee.
-
2011-2013 Chairman, The Biochemical Society
-
2010 Co-chair, ESF-EMBO conference on Molecular Perspectives on Protein-Protein Interactions, Sant Feliu de Guixols, Spain
-
2010-2014 Faculty 1000 reviewer-Protein Chemistry & Proteomics
-
2010-2011 Editorial Advisory Board, Molecular Microbiology
-
2008 External assessor, Department of Biochemistry, University of Bristol
-
2008-2009 EPSRC Chemistry International Review Steering Committee
-
2008-2010 Vice-chair, Biochemical Society
-
2008 Chair, 2nd international conference on Molecular Perspectives on Protein-Protein Interactions, Dubrovnik, Croatia
-
2008 Co-chair, Gordon Research Conference on Biomolecular Interactions & Methods, Ventura, CA
-
2007-2010 Wellcome Trust Basic Science Interview committee
-
2007 BBSRC REI panel
-
2006- 2008 External examiner for BSc (Hons) Degree Programmes in Biochemistry, Molecular Biology and Molecular Genetics, University of Dundee
-
2006 - 2008 External examiner for 1st year MRes of Wellcome Trust 4 year PhD programme at the University of Glasgow
-
2006 International vice-chair for Gordon Research Conference on Reversible Associations in Structural & Molecular Biology, Ventura, CA
-
2005 Member of organising scientific committee for the Katzir conference on Molecular Perspectives on Protein-Protein Interactions, Eilat, Israel
-
2005 Contributor to Faculty of 1000 Biology - Protein Chemistry & Proteomics
-
2005 BBSRC Integrative & Systems Biology Strategy panel
-
2005 External RAE adviser to the School of Biological Sciences, University of East Anglia
-
2002 - 2006 Biochemistry and Cell Biology panel of the BBSRC
-
2004-2007 External ‘critical friend’ for RAE08 Department of Biochemistry, University of Sussex
-
2002 Co-opted panel member for Wellcome Trust Functional Genomics Development Initiative
-
1995 - 1997 Molecular Enzymology Group of the Biochemical Society
-
1994 - 1997 Editorial Board of the Biochemical Journal
-
1994 - 1997 Molecular and Cell Panel of The Wellcome Trust
-
Member of the Biophysical Society
-
Member of the Biochemical Society
Recent invited lectures
2018
- Honorary lecture, 80th birthday celebrations of Prof Volkmar Braun, University of Tübingen, Germany
- EMBO members meeting, Heidelberg, Germany
2017
- Hull-York Medical School, University of Hull
- Oxford University Biochemical Society, Department of Biochemistry, Oxford
- Department of Microbiology & Immunobiology, Harvard Medical School, Boston, USA
- 7th Congress of European Microbiologists, FEMS 2017, Valencia, Spain
- Inaugural Glasgow Microbiology Collective meeting, University of Strathclyde
- Protein Folding, Evolution and Interactions Symposium, University of Cambridge
- New Horizons in Membrane Transport & Communication, CRC807, Goethe University, Frankfurt, Germany
- Faculty of Biology & Chemistry, University of Osnabrȕck, Germany
- 6th International conference on Molecular Perspectives on Protein-Protein Interactions, Eilat, Israel.
2016
- Keynote, Mechanisms of bacterial toxin delivery symposium, UC Santa Barbara, USA
- Department of Infectious Diseases, Genetech, USA
- University of Southern Florida, USA
- Protein secretion in bacteria, Zing Conference, St. Petersburg, FL, USA
2015
- 40th Lourne conference on Protein Function and Sctructure, Lourne, Australia
- i3 Institute University of Technology Sydney, Australia
- School of Chemistry and Molecular Biosciences, University of Brisbane, Australia
- Department of Biological Chemistry, Weizmann Institute of Science, Israel
- 5th International conference on Molecular Perspectives on Protein-Protein interactions, Toronto, Canada
- Gordon Research Conference on Proteins, Holderness College, NH, USA
- Leibniz Institute of Molecular Pharmacology, FMP Berlin, Germany
2014
- Gordon Research Conference on Bacterial Cell Surfaces, Mount Snow, VT, USA
- Gordon Research Conference on Intrinsically Disordered Proteins, Stonehill College, MA, USA
- Protein Society, San Diego, California, USA
2013
- Randall Institute, King’s College London
- Microbes in Norwich symposium & JIC open seminar, John Innes Centre, Norwich
- Department of Biochemistry annual recess, Oxford
- ESF-EMBO conference on Molecular Perspectives on Protein-Protein Interactions, Pultusk, Poland
- Marie Curie TRANSPOL network summer school, Pultusk, Poland
- Keynote speaker, Inaugural South West Doctoral Training Partnership Conference
- 10th annual EIMID meeting, Couvent Royal de Saint-Maximin, France
- AFMB, FRISBI workshop, Marseille, France
- School of Life Sciences, University of Warwick
2012
- Biochemical Society Focus Meeting, How bugs kill bugs: Progress & challenges in bacteriocin research. University of Nottingham, School of Biological Sciences, University of Canterbury, Christchurch, New Zealand
- Biochemical Society lecture, Queenstown Molecular Biology 2012 meeting, New Zealand
- Young Microbiologists Symposium, University College Cork, Ireland
2011
- 103rd meeting of the Scottish Protein Structure Group, University of Edinburgh
- 50th Anniversary of the University of Leicester Biochemistry Department
- Society of General Microbiology, Cell Envelope: Architecture, University of York
- Institute for Cell & Molecular Biosciences, University of Newcastle
- Biochemical Society sponsored seminar, John Innes Centre, Norwich
- British Crystallography Association, winter meeting, Harwell, Didcot
2010
- Gordon Research Conference on Ligand Recognition & Molecular Gating, Il Ciocco, Lucca, Italy
- Speaker and Session chair, Advances in Biophysical Methods, Gordon Research Conference on Biomolecular Interactions & Methods, Galveston, Tx, USA
- Division of Infection and Immunity and Wellcome Centre for Molecular Parasitology, University of Glasgow
2009
- Department of Pathology, University of Cambridge
- Department of Crystallography, Birkbeck College, University of London
- Conway Institute of Biomolecular and Biomedical Research, University College Dublin
- National Institute of Medical Research, Mill Hill, London
- Division of Molecular Biosciences, Imperial College, London
- Dahlem Lecture, Max Planck Institute for Molecular Genetics, Berlin
- Division of Molecular Microbiology, University of Dundee
- Faculty of Biological Sciences, University of Leeds
- School of Biological Sciences, Queen’s University Belfast
2008
- School of Biological Sciences, University of East Anglia, Norwich
- Department of Molecular Biology & Biotechnology, University of Sheffield
- 2nd International Conference on Molecular Perspectives on Protein-Protein Interactions, Dubrovnik, Croatia
- ESF-EMBO Bacterial Networks meeting (BacNet08), Barcelona, Spain
- Strategies for intrinsically unfolded proteins, International Conference on Structural Genomics, Oxford
- Inaugural lecture, University of York
- Boston Biomedical Research Institute, Boston, MA, USA
- NIDDK, NIH, Bethesda, USA
2007
- Department of Biochemistry, University of Bristol
- Department of Biochemistry, University of Birmingham
- Laboratory of Molecular Biology, Cambridge
- Society for General Microbiology, Cells and Cell Surfaces, University of Manchester
- Department of Chemistry, University of Edinburgh
- Department of Biology, University of York
- 6th European Biophysics Congress, session on Disordered and Aggregated Proteins, Imperial College London
- Institute of Chemistry, University of Leiden, The Netherlands
- School of Biosciences, University of Nottingham
2006
- Department of Biosciences, University of Kent
- Department of Protein Engineering, Genentech, South San Francisco, USA
- FASEB Summer Research Conference, Nucleic Acid Enzymes, Vermont, USA
- 1st UK Bacterial Cell Wall Biosynthesis Network meeting, University of Warwick
- ProSA Marie-Curie Network workshop, MPI, Berlin, Germany
- John Innes Centre, Norwich Research Park, Norwich
2005
- Biomolecular Dynamics & Interactions Symposium, Bio21 Institute, University of Melbourne, Australia
- 30th Annual Lorne Conference on Protein Structure & Function, Phillip Island, Australia
- School of Molecular and Microbial Sciences, University of Brisbane, Australia
- Bioinformatics Institute, Biopolis, Singapore
- York Biology Cell Signalling minisymposium
- Head of Department Seminar series, York
- Speaker and session chair at the Katzir conference on Molecular Perspectives on Protein-Protein Interactions, Eilat, Israel
2004
- Speaker and session chair, Gordon Research Conference on Reversible Associations in Structural & Molecular Biology, Ventura, CA
- Department of Biomolecular Sciences, University of St. Andrews
- Protein Structure & Folding minisymposium, Faculties of Pharmacy & Chemistry, University of Utrecht, The Netherlands
- 5th International Conference on ‘Zinc Signals’, University of Aarhus, Denmark
- Invited speaker at EMBO Workshop on Transient kinetics applied to biological macromolecules, University of Kent, Canterbury
2003
- Department of Biological Sciences, University of Southampton
- Department of Biochemistry, University of Glasgow
- Society of General Microbiology sponsored seminar, Department of Microbiology, Moyne Institute for Preventative Medicine, Trinity College, Dublin
- Department of Biological Sciences, University of Hull
2002
- 40th Anniversary Celebrations, Department of Biochemistry, University of Leicester
2001
- Norwich Research Park Symposium on ‘Proteins: Cells, Systems & Applications’
- Department of Biochemistry and Genetics, University of Newcastle
- Cambridge Centre for Molecular Recognition, University of Cambridge
- Biochemical Society (Bristol), symposium on Molecular Communications
- Departments of Chemistry and Biology Joint Seminar, University of York
- Department of Microbiology & Immunology, Queen’s University of Belfast
- Department of Biosciences, University of Birmingham
2000
- Speaker and Session Chair, Gordon Research Conference on Bacterial Cell Surfaces, Colby-Sawyer College, NH
- Plenary speaker, Macromolecules in Chemistry and Biology symposium, University of Lancaster
- Bijvoet Centre, University of Utrecht, The Netherlands
Postdoctoral Research Associates
- Nick Housden
- Gideon Mamou (EMBO LTF)
- Soumik Basu (Royal Society Newton International Fellow)
- Ruth Cohen-Khait
- Nathalie Reichmann
- Melissa Webby
- Sandip Kumar
- Joanna Szczepaniak
Graduate Students
Research Support
Part II
Current Collaborators
- Professor Martin Maiden, Department of Zoology, University of Oxford
- Professor C.V. Robinson FRS, Department of Chemistry, University of Oxford.
- Professor Hagen Bayley FRS, Chemical Biology, Department of Chemistry, University of Oxford.
- Professor Helen Saibil FRS, Department of Biological Sciences and ISMB, Birkbeck College
- Professor Mark Sansom, Department of Biochemistry, University of Oxford.
- Professor Jacob Piehler, Division of Biophysics, Universität Osnabrück.
- Dr Christoph Baumann, Department of Biology, University of York
- Dr Daniel Walker, Institute of Infection, Immunity and Inflammation, University of Glasgow
- Dr Julian Parkhill FRS, The Sanger Institute, Cambridge
- Professor Peter Taylor, School of Pharmacy, UCL
Post Doctoral Research Associates |
PhD students |
|
Visitors
Technicians
|
|
|
|
Masters students
|
||
Part II & Summer Students
|
* Have established their own labs

The 5th International Conference on Molecular Perspectives on Protein-Protein Interactions
29 May - 2 June 2015

Lorne Proteins 2015
The 40th Lorne Conference on Protein Structure and Function
8-12 Feb 2015
Mantra Lorne, Victoria, Australia
Intrinsically disordered proteins
Gordon Research Conferences
Understanding Intrinsically Disordered Regions (IDRs) at Different Scales: From Single Molecules to Complex Systems
6-11 July 2014
Stonehill College Easton, MA
Bacterial cell surfaces
Gordon Research Conferences
Building, Splitting, and Traversing the Cell Surface
22-27 June 2014
Mount Snow West Dover, VT

The Protein Society Symposium
The 28th Annual Symposium of the Protein society
27-30 July 2013
San Diego United States

ESF-EMBO Symposium
Molecular Perspectives On Protein-Protein Interactions
25-30 May 2013
Polonia Castle in Pultusk, Poland
Chaired by: Prof. Marcellus Ubbink, University of Leiden
Co-chaired by: Gideon Schreiber, Weizmann Institute of Science, Colin Kleanthous, Oxford University








