Breakthrough for Optogenetics
The structure of a protein used in over 1000 labs worldwide has been resolved for the first time by an international team lead by the Watts lab
The photoreceptor AR3 is expressed by Halorubrum sodomense, an organism that grows in the Dead Sea, but is best known for its very widespread (>1000 labs) applications in optogenetics experiments, in which it is used to silence individual neurons and to detect changes in cell membrane voltage. These new structures give very high-resolution details (at an unprecedented 1.07Å resolution) that open the way for the development of new tools and methodologies in the fields of neuroscience, cell biology, biotechnology and beyond.
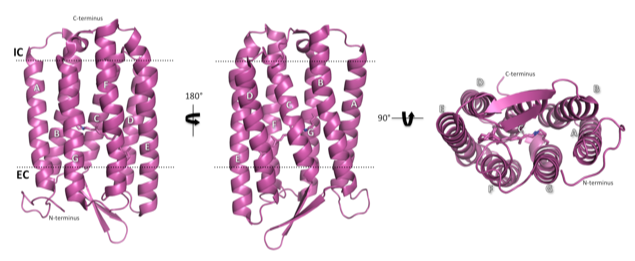
The structure of light-adapted (LA) AR3 (PDB accession code: 6S6C). The seven transmembrane helices are labelled A-G.
Working with Dr Isabel Moraes (National Physical Laboratory) and several experts at Diamond Light Source, Dr Juan Bada Juarez and Dr Peter Judge were able to crystallise AR3 in two distinct conformations - the ground state, in which the protein is configured to transport one H+ ion across the cell membrane for each photon absorbed and a desensitized state which it adopts in the prolonged absence of light. By comparing the two structures, they were able to identify the remodelling that takes place in the internal hydrogen bonding networks to enable the photoreceptor to switch between the two states.
Crystals of AR3 observed under polarised light
The work, led by Tony Watts from the Biochemistry Department, also involved multi-national research groups from France and Israel. Together, the team characterised the protein, which has resisted many previous efforts at crystallisation. Techniques including native mass spectrometry (with Carol Robinson's group in Oxford), QMMM calculations (with Igor Schapiro 's group in Jerusalem) and AFM (with Pierre-Emmanuel Milhiet's group in Montpellier) have given new insights into the mechanism of this highly efficient proton pump, and the importance of water and hydrogen bonding in receptor desensitisation.
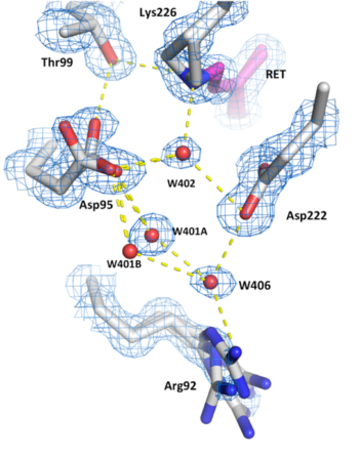
Hydrogen bond networks inside the desensitised state of AR3
Corresponding author Prof. Anthony Watts from Oxford University said:
"The superb resolution that we have achieved for these AR3 structures, 1.07 Å for the ground state, is among the highest for a wild-type membrane protein deposited to date in the Protein Data Bank. This quality allows us to visualise directly the complex distribution of water molecules inside the receptor and describe the functional significance of the intricate networks of hydrogen bonds that they form, something that is of importance in many biomolecules - not just photoreceptors."
Reference
- Bada Juarez, J.F., Judge P.J. et al. Structures of the archaerhodopsin-3 transporter reveal that disordering of internal water networks underpins receptor sensitization Nature Communications (2021) DOI: 10.1038/s41467-020-20596-0
Deposited structures
- 6S6C - Ground state structure of Archaerhodopsin-3 at 100K - 1.07 Å resolution
- 6GUX - Dark-adapted structure of Archaerhodopsin-3 at 100K- 1.3 Å resolution
Anthony Watts
26th January 2021




