Microbiology and Systems Biology
We aim to uncover how eukaryotes and bacteria carry out fundamental processes in order to survive. Our interests span the molecular mechanisms of proteins and their interaction networks all the way to cell-cell communication in microbes. Specific areas of interest include bacterial DNA repair, signalling, motility, transport and intercellular competition and yeast cell cycle and metabolic control. We explain these processes using a combination of experimental, modelling, computational and mathematical tools, at scales ranging from atomic resolution to whole cells and cell populations. We also develop synthetic biology approaches to enhance and exploit biological functions.
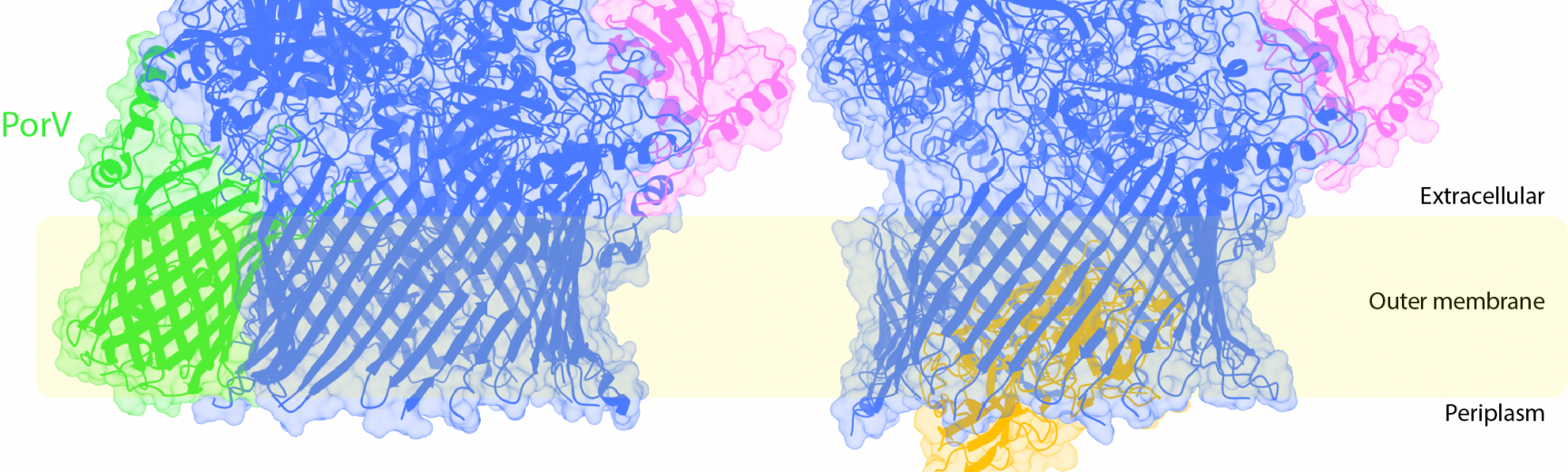
Our research centres on microbes and spans areas such as the molecular mechanisms of protein machines, bacterial pathogenesis, cellular homeostasis control and cell population dynamics. Ben Berks’s lab studies complex nanomachines that allow selective movement of proteins and DNA across the different barriers of the bacterial cell envelope and how this movement is energized. Stephan Uphoff’s lab investigates the mechanisms underlying bacterial adaptation, DNA repair, and mutagenesis, using novel microscopy techniques developed to visualise these processes within cells. Syma Khalid's lab uses molecular dynamics and other computational methods to predict the movements of proteins, lipids, sugars and other macromolecules. Mark Roberts focusses on how we teach the practical side of biochemistry, alongside how we teach data analysis and interpretation. Tobias Warnecke studies the evolution of chromatin from a comparative perspective, with a particular focus on less-studied organisms such as archaea. Georgina Benn's goal is to understand how the outer membrane is organised, how its mechanical properties are distributed, and how it is deformed during cell division. Dmitry Ghilarov's group works to reveal fundamental principles of how bacterial molecular machines use energy to manipulate three-dimensional structure of peptides and nucleic acids.